Template
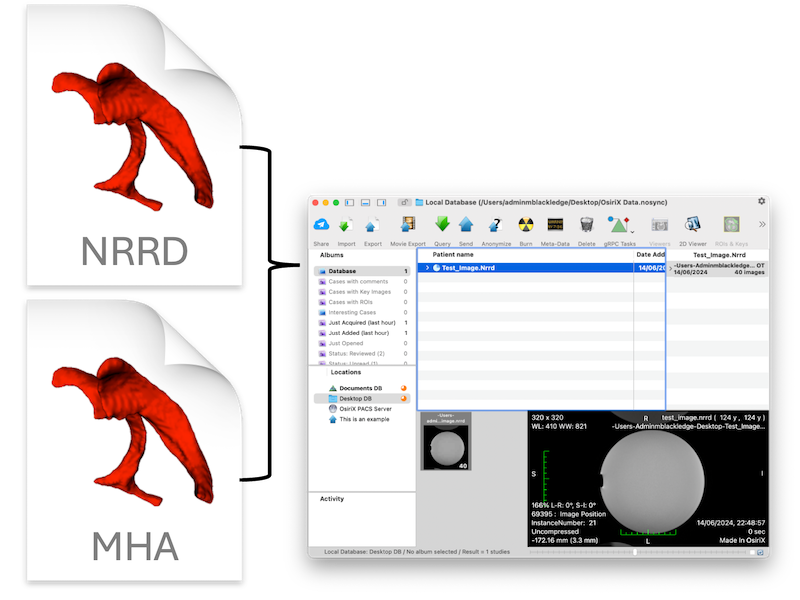
A script for importing and visualizing ITK images (e.g. nrrd, mhd/mha, nifti) within OsiriX.
Categories:
Authors
| Author(s) | Matthew Blackledge1 |
| Institution(s) | 1The Institute of Cancer Research |
| Contact | matthew.blackledge@icr.ac.uk |
Datasets
Any data.
Details
This script will read an ITK image format, create basic Dicom files, and load them into OsiriX for viewing.
Note that all created Dicom files will have SOP class "Secondary Capture Image" by default. Please change the relevant
line in the code ds.SOPClassUID = "1.2.840.10008.5.1.4.1.1.7" to the necessary image type if required.
Each ITK image will have a unique series and study UID.
Expected Outcome
Should load the ITK image into the OsiriX database for visualization.
Script
""" A script for reading and loading ITK images into OsiriX """
import os
import sys
import datetime
import uuid
import shutil
from time import sleep
import osirix
import SimpleITK as sitk
import numpy as np
import pydicom
from pydicom.dataset import Dataset, FileDataset, FileMetaDataset
from pydicom.uid import generate_uid, ExplicitVRLittleEndian, UID
from PyQt5.QtWidgets import QFileDialog, QApplication
def select_file():
""" A simple widget for selecting a ITK file.
"""
QApplication(sys.argv)
options = QFileDialog.Options()
options |= QFileDialog.ReadOnly
file_name, _ = QFileDialog.getOpenFileName(None, "Select File", "", "ITK Files (*.nrrd *.mha *.mhd *.nifti)", options=options)
if file_name:
return file_name
else:
return None
def image_to_patient_coordinates(ijk, direction, spacing, origin):
""" Transform image to patient coordinates (ijk -> xyz)
"""
trans = np.array([[direction[0] * spacing[0], direction[3] * spacing[1], direction[6] * spacing[2], origin[0]],
[direction[1] * spacing[0], direction[4] * spacing[1], direction[7] * spacing[2], origin[1]],
[direction[2] * spacing[0], direction[5] * spacing[1], direction[8] * spacing[2], origin[2]],
[0, 0, 0, 1]])
xyz = np.dot(trans, np.r_[ijk, 1])[0:3]
return xyz
def scale_array(array, bits = 12):
""" Used to scale an input array in the range 0 to 2**bits
"""
window_max = float(2**bits)
min, max = np.nanmin(array), np.nanmax(array)
rescale_slope = (max - min) / window_max
rescale_intercept = min
scaled_array = (array - rescale_intercept) / rescale_slope
return scaled_array.astype("int"), rescale_intercept, rescale_slope
def generate_accession_number():
""" Generate a unique ID to use as an accession number
"""
accession_number = str(uuid.uuid4())
short_accession_number = accession_number[:16]
return short_accession_number
def create_dicom_series_from_itk(itk_file, output_dir):
""" Create and save Dicom files for an ITK file (e.g. nrrd, nifti)
"""
# Read the file
image = sitk.ReadImage(itk_file)
# Get the numpy array from the image
image_array = sitk.GetArrayFromImage(image).astype("float")
# Get image metadata
origin = image.GetOrigin()
spacing = image.GetSpacing()
direction = image.GetDirection()
study_uid = pydicom.uid.generate_uid()
study_id = str(np.random.randint(10000, 99999, size = 1))
series_uid = pydicom.uid.generate_uid()
series_number = np.random.randint(10000, 99999, size = 1)
# Ensure output directory exists
os.makedirs(output_dir, exist_ok=True)
# Iterate through each slice and create a DICOM file
for i in range(image_array.shape[0]):
# Scale the input array
bits = 12
slice_array = image_array[i, :, :]
slice_array, rintercept, rslope = scale_array(slice_array, bits=bits)
slice_array = slice_array.astype(f"uint16")
# Create a new DICOM dataset
file_meta = FileMetaDataset()
file_meta.MediaStorageSOPClassUID = UID("1.2.840.10008.5.1.4.1.1.7")
file_meta.MediaStorageSOPInstanceUID = pydicom.uid.generate_uid()
file_meta.TransferSyntaxUID = pydicom.uid.ExplicitVRLittleEndian
file_meta.ImplementationClassUID = pydicom.uid.PYDICOM_IMPLEMENTATION_UID
# Calculate the File Meta Information Group Length
file_meta.FileMetaInformationGroupLength = 132 + len(file_meta)
# Create the FileDataset instance (initially no data elements, but file_meta supplied)
ds = FileDataset(f'slice_{i:04d}.dcm',
{},
file_meta=file_meta,
preamble=b"\0" * 128)
# Set creation date/time
dt = datetime.datetime.now()
date_str = dt.strftime('%Y%m%d')
time_str = dt.strftime('%H%M%S.%f')
ds.ContentDate = date_str
ds.StudyDate = date_str
ds.SeriesDate = date_str
ds.AcquisitionDate = date_str
ds.InstanceCreationDate = date_str
ds.ContentTime = time_str
ds.StudyTime = time_str
ds.SeriesTime = time_str
ds.AcquisitionTime = time_str
ds.InstanceCreationTime = time_str
# Set the transfer syntax
ds.is_little_endian = True
ds.is_implicit_VR = False
# Add necessary metadata
ds.SpecificCharacterSet = "ISO_IR 100"
ds.ImageType = ['DERIVED']
ds.SOPClassUID = "1.2.840.10008.5.1.4.1.1.7"
ds.SOPInstanceUID = file_meta.MediaStorageSOPInstanceUID
ds.StudyInstanceUID = study_uid
ds.SeriesInstanceUID = series_uid
ds.SeriesNumber = series_number
ds.StudyID = study_id
ds.InstanceNumber = i+1
ds.SeriesDescription = itk_file
ds.PatientName = os.path.basename(itk_file)
ds.PatientID = os.path.basename(itk_file)
ds.PatientBirthDate = "19000101"
ds.PatientSex = "O"
ds.PatientWeight = ""
ds.ReferringPhysicianName = ""
ds.ImagePositionPatient = image_to_patient_coordinates([0, 0, i],
direction,
spacing,
origin).tolist()
ds.ImageOrientationPatient = [d for d in direction[:6]]
ds.PixelSpacing = [spacing[0], spacing[1]]
ds.SliceThickness = spacing[2]
ds.SliceInterval = spacing[2]
ds.Modality = "OT" # Other
ds.Manufacturer = "OSIRIXGRPC"
ds.Rows, ds.Columns = slice_array.shape
ds.PhotometricInterpretation = "MONOCHROME2"
ds.SamplesPerPixel = 1
ds.BitsAllocated = 16
ds.BitsStored = bits
ds.HighBit = bits-1
ds.RescaleIntercept = rintercept
ds.RescaleSlope = rslope
ds.RescaleType = "US"
ds.ConversionType = "SYN"
ds.AccessionNumber = generate_accession_number()
ds.PixelRepresentation = 0 # Unsigned integers
# Set pixel data
ds.PixelData = slice_array.tobytes()
# Ensure that the File Meta Information Group Length is calculated and written
pydicom.dataset.validate_file_meta(ds.file_meta)
# Save the DICOM file
output_file = os.path.join(output_dir, f"slice_{i:04d}.dcm")
ds.save_as(output_file)
def run_script():
""" Run the pyOsiriX script
"""
selected_file = select_file()
if not selected_file:
return # User clicked cancel
# Temporary storage
temp_out = ".itk2dicom_temp"
os.makedirs(temp_out, exist_ok=True)
# Create the Dicom files
create_dicom_series_from_itk(selected_file, temp_out)
# Load into the database
bc = osirix.current_browser()
current_directory = os.getcwd()
file_list = [os.path.join(current_directory, temp_out, fl) for fl in os.listdir(temp_out)]
bc.copy_files_into_database(file_list)
# Wait until loaded (1s should be enough) and then delete
sleep(1)
shutil.rmtree(temp_out)
if __name__ == "__main__":
run_script()
